Furthermore, the endogenously-generated Th17 response is qualitatively distinct from that generated as a result of adoptive transfer of in vitro polarized Th17 cells, both in its sensitivity to GC and its requirement for IL-1R signaling. These data demonstrate that the Th17 response generated endogenously is substantially different than the response acquired upon adaptive transfer of in vitro polarized Th17 cells and OVA challenge, suggesting that in some instances, the outcomes of an adoptive transfer study may not be generalizable to other models of allergic airway disease. Clarification of where these models fit in terms of clinical asthma demands robust comparative analyses. As exemplified by our findings, the functional significance of the Th17 response in promoting allergic airway disease is determined by the conditions in which it was generated, ranging from protective to pathogenic. Given the heterogeneity among patients with clinical asthma, the AbMole Nitisinone contribution of a single type of immune response may also vary among patients, which should be considered during pharmacologic development and clinical testing. One of the main fields in biological researches is to reveal biological regulatory networks that control different functions. In the past, molecular interactions have been established at a rather slow pace. For example, it took more than a decade from the discovery of the well-known tumor suppressor gene p53 to the establishment of its regulatory feedback loop with the protein MDM2. This situation has been qualitatively changed thanks to the 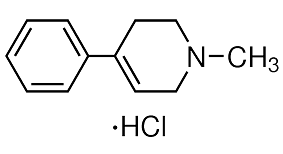 development of different new biotechnologies, especially microarray expression experiments. Accordingly, theoretical systems biology has provided several algorithms for the deduction of regulatory interactions from experimental data. These algorithms can be employed to effectively reconstruct biological regulatory networks. One of the successful network reconstruction AbMole Nitroprusside disodium dihydrate methods is reverse engineering approach, which has been advancing very rapidly in recent years. At present, there are mainly four types of the reverse engineering algorithms: correlation-based methods, information-theoretic methods, Bayesian network predictions, and methods based on dynamic models. In this paper, we apply the simplest method, the correlation-based Boolean reverse engineering, to discuss algorithms for the optimization of experiments in network construction. In this paper, we discuss a reverse engineering approach to reconstruct biological regulatory network from experiments. Instead of using real data from experiments, we applied three well-known regulatory networks as tests to generate the trajectories of the networks. We focus on how to design the experiment to get the most useful information from data of evolution trajectory. For this purpose, we propose three methods, namely, maximum distance method, trajectory entropy method, and sampling method, to derive the optimal initial conditions for experiments. The performance of these methods is discussed and evaluated by comparing the reconstructed networks with the original ones in three known systems. We also test our methods under more realistic circumstance when the number of nodes that can be perturbed is limited. From these analyses we conclude that the sampling method is the best among the three methods. Two issues are called for further investigation. The calculation cost of entropy method increases very rapidly as the node number increase; it becomes very time-consuming when the node number is large. In contrast, the calculation time of max-distance and sampling methods increase very slowly with the increase of the node number of network.
development of different new biotechnologies, especially microarray expression experiments. Accordingly, theoretical systems biology has provided several algorithms for the deduction of regulatory interactions from experimental data. These algorithms can be employed to effectively reconstruct biological regulatory networks. One of the successful network reconstruction AbMole Nitroprusside disodium dihydrate methods is reverse engineering approach, which has been advancing very rapidly in recent years. At present, there are mainly four types of the reverse engineering algorithms: correlation-based methods, information-theoretic methods, Bayesian network predictions, and methods based on dynamic models. In this paper, we apply the simplest method, the correlation-based Boolean reverse engineering, to discuss algorithms for the optimization of experiments in network construction. In this paper, we discuss a reverse engineering approach to reconstruct biological regulatory network from experiments. Instead of using real data from experiments, we applied three well-known regulatory networks as tests to generate the trajectories of the networks. We focus on how to design the experiment to get the most useful information from data of evolution trajectory. For this purpose, we propose three methods, namely, maximum distance method, trajectory entropy method, and sampling method, to derive the optimal initial conditions for experiments. The performance of these methods is discussed and evaluated by comparing the reconstructed networks with the original ones in three known systems. We also test our methods under more realistic circumstance when the number of nodes that can be perturbed is limited. From these analyses we conclude that the sampling method is the best among the three methods. Two issues are called for further investigation. The calculation cost of entropy method increases very rapidly as the node number increase; it becomes very time-consuming when the node number is large. In contrast, the calculation time of max-distance and sampling methods increase very slowly with the increase of the node number of network.