Patients with DV infection show various clinical symptoms that range from no significant illness or mild fever to life-threatening Dengue hemorrhagic fever and Dengue shock syndrome. Currently, only supportive treatments are available. Although considerable research has been AZD2281 directed towards the development of a safe and effective DV vaccine since the mid-20th century, there are no approved commercial products available. Therefore, to combat DV and other related viral diseases, it is advisable to develop novel strategies for discovering new antiviral agents. Recent progress in the biology has brought with it many protein structures for virtual screening as drug targets. However, without a previously validated target site on the targeted protein as a reference point, the number of lead candidates obtained from this type of screening is very large. Cellular toxicity further complicates biological activity assays as well. Therefore, the utilization of VS is somewhat hindered by the processes that follow, namely, the labor-intense, time-consuming verification process and the toxicity assays required for processing large amounts of lead candidates. Here, in an attempt to devise a less resource-demanding screening process, we have focused on computational approaches that are solely based on the structures of a designated region of the target protein. Then, we performed VS on a set of medical compounds because we recognized that using medical compounds could potentially minimize cellular toxicity. To reduce the number of lead candidates, we further refined the VS output by structural clustering for the identification of novel structural characteristics. Compounds with novel structures were then subjected to a biological assay to validate their activities. In summary, we sacrificed the diversity of leads in exchange for the efficiency of screening. The DV envelope protein is 495 amino acids in length, forms oligomers, and, along with the M protein, constitutes most of the accessible virion surface that is covered by the envelope membrane. The E protein is responsible for activating “membrane fusion”, the central molecular event during the entry of enveloped RNA viruses into host cells. 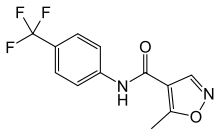 The Dengue virus enters a host cell when the E protein binds to the virus receptor on the host cell surface and activates its conformational rearrangement, causing the E protein in its dimeric pre-fusion form to transform into a trimeric post-fusion structure. This essentially irreversible conformational change induces the fusion between the viral envelope membrane and the host cell membrane, allowing entry to be completed. In short, the DV E protein mediates host cell binding and is essential for infection via a conformationinduced membrane fusion event between the host cell and the virion. In addition, it is also the primary antigen that induces protective immunity and the major antigen for virus neutralization. The crystal structures of the E protein of DV type 2 in both the presence and absence of a bound ligand were deposited in the Protein Data Bank {PDB codes 1oke and 1ok8, respectively; Figure 1). The key difference between these two structures is a local rearrangement of the “kl” b-hairpin and the concomitant opening up of a hydrophobic pocket for ligand binding. For example, the detergent noctyl-b-D-glucoside can occupy this pocket. Mutations that affect the pH threshold for membrane fusion have also been mapped to this hydrophobic pocket. Therefore, Modis et al. proposed that this pocket was a hinge point in the fusionactivating conformational change. This concept made the utilization of structure-based VS to identify inhibitors of DV infection plausible. Therefore, in this study, a well-developed HhAntag691 docking tool, GEMDOCK, was utilized to perform VS on the Comprehensive Medicinal Chemistry databasefor substances thatcoulddock in this hydrophobic pocket of E proteins.
The Dengue virus enters a host cell when the E protein binds to the virus receptor on the host cell surface and activates its conformational rearrangement, causing the E protein in its dimeric pre-fusion form to transform into a trimeric post-fusion structure. This essentially irreversible conformational change induces the fusion between the viral envelope membrane and the host cell membrane, allowing entry to be completed. In short, the DV E protein mediates host cell binding and is essential for infection via a conformationinduced membrane fusion event between the host cell and the virion. In addition, it is also the primary antigen that induces protective immunity and the major antigen for virus neutralization. The crystal structures of the E protein of DV type 2 in both the presence and absence of a bound ligand were deposited in the Protein Data Bank {PDB codes 1oke and 1ok8, respectively; Figure 1). The key difference between these two structures is a local rearrangement of the “kl” b-hairpin and the concomitant opening up of a hydrophobic pocket for ligand binding. For example, the detergent noctyl-b-D-glucoside can occupy this pocket. Mutations that affect the pH threshold for membrane fusion have also been mapped to this hydrophobic pocket. Therefore, Modis et al. proposed that this pocket was a hinge point in the fusionactivating conformational change. This concept made the utilization of structure-based VS to identify inhibitors of DV infection plausible. Therefore, in this study, a well-developed HhAntag691 docking tool, GEMDOCK, was utilized to perform VS on the Comprehensive Medicinal Chemistry databasefor substances thatcoulddock in this hydrophobic pocket of E proteins.